Calibration
HydroCNHS is equipped with a genetic algorithm package powered by Distributed Evolutionary Algorithms in Python (DEAP), which can be used for calibration in parallel. HydroCNHS is unique in that aspect that both the parameters of the hydrological model and user-defined ABM can be calibrated simultaneously as long as they are defined in the model configuration file (.yaml). Furthermore, users are allowed to assign initial guesses to the algorithm.
Calibration with the genetic algorithm (GA)
An evaluation function, a calibration input dictionary, and a GA configuration dictionary are required to use the GA calibration module supported by the HydroCNHS.
Evaluation function
Assuming we want to maximize \(y=-x_1^2+5x_1-x_2\) with \(x_1,x_2 \in [-5,5]\), we can design the evaluation function as:
import os
import HydroCNHS
import HydroCNHS.calibration as cali
prj_path, this_filename = os.path.split(__file__)
def evaluation(individual, info):
x = individual
fitness = -x[0]**2 + 5*x[0] - x[1]
return (fitness,)
cali_wd, current_generation, ith_individual, formatter, _ = info
name = "{}-{}".format(current_generation, ith_individual)
##### individual -> model
# Convert 1D array to a list of dataframes.
df_list = cali.Convertor.to_df_list(individual, formatter)
# Feed dataframes in df_list to model dictionary.
The evaluation must have “individual” and “info” arguments. Also, the return fitness value has to be of tuple format, e.g., (fitness,). The “info” contains additional information for users to design a more complex evaluation, as shown in “Build a hydrological model” and “Integrate an ABM” sections. This “info” variable contains information, including 1. working directory to the folder of this calibration experiment (cali_wd), 2. current generation of the GA run (current_generation), 3. index of the individual in the current generation (ith_individual), 4. formatter (formatter) for a converter, and 5. NumPy random number generator (rn_gen).
cali_wd, current_generation, ith_individual, formatter, rn_gen = info
cali_wd, current_generation, and ith_individual can be used to create sub-folders or output files for each evaluation. This is especially useful for calibrating a large model that requires a long simulation time. The formatter contains information to convert 1-D array individuals back to the original parameter format (e.g., a list of DataFrames) through a converter object. We will introduce the Converter in the next section. rn_gen is recommended to be used to generate random numbers (e.g., input to a HydroCNHS object like model = HydroCNHS.Model(model, name, rn_gen)). By applying rn_gen, HydroCNHS can guarantee the reproducibility of the numerical experiment with a given random seed.
Calibration input dictionary
A calibration input dictionary contains three keys, including par_name, par_bound, and wd. par_name is a list of the parameter names, par_bound is a list of parameter bounds, and wd is the working directory for the calibration experiment. The calibration input dictionary for this example is shown below.
# Calibration inputs
cali.get_inputs_template() # print an input template.
inputs = {'par_name': ['x1', 'x2'],
'par_bound': [[-5, 5], [-5, 5]],
'wd': 'working directory'}
inputs["wd"] = prj_path
Note that users can use a converter to generate the calibration input dictionary automatically.
GA configuration dictionary
A GA configuration dictionary contains many control options. Please see the following code for the explanation of each option.
# GA configuration
cali.get_config_template()
config = {'min_or_max': 'max', # maximize or minimize the evaluation function.
'pop_size': 100, # Size of the population.
'num_ellite': 1, # Number of ellites.
'prob_cross': 0.5, # Crossover probability for uniform crossover operator.
'prob_mut': 0.15, # Mutation probability of each parameter.
'stochastic': False, # Is the evaluation stochastic?
'max_gen': 100, # Maximum generation number.
'sampling_method': 'LHC', # Sampling method for the initial population.
'drop_record': False, # Whether to drop historical records to save space.
'paral_cores': -1, # Number of parallel cores. -1 means all available cores.
'paral_verbose': 1, # Higher value will output more console messages.
'auto_save': True, # If true, users may continue the run later on by loading the auto-save file.
'print_level': 1, # Control the number of generations before the printing summary of GA run.
'plot': True} # Plot to time series of the best fitnesses over a generation.
Finally, we can run the GA calibration.
# Run GA
rn_gen = HydroCNHS.create_rn_gen(seed=3)
ga = cali.GA_DEAP(evaluation, rn_gen)
ga.set(inputs, config, formatter=None, name="Cali_example")
ga.run()
ga.solution
# Out[0]: array([ 2.47745344, -4.96991833])
After the GA terminated, we got the solution \(x_1=2.4775\) and \(x_2=-4.9699\), in which the theoretical values are 2.5 and -5 for \(x_1\) and \(x_2\), respectively. The fitness plot (Fig. 7) and auto-saved file (GA_auto_save.pickle) will be stored in the GA working directory. This GA_auto_save.pickle can be loaded into the GA object and continue the calibration when algorithms encounter a sudden breakdown or continue the run with a larger “max_gen,” as shown below.
# Continue the run with larger "max_gen"
ga = cali.GA_DEAP(evaluation, rn_gen)
ga.load(os.path.join(prj_path, "Cali_example", "GA_auto_save.pickle"),
max_gen=120)
ga.run()
# =====Generation 120=====
# Elapsed time 00:00:05
# Min -6.69464
# Max 11.21948
# Avg 10.99626
# Std 1.77931
# GA done!
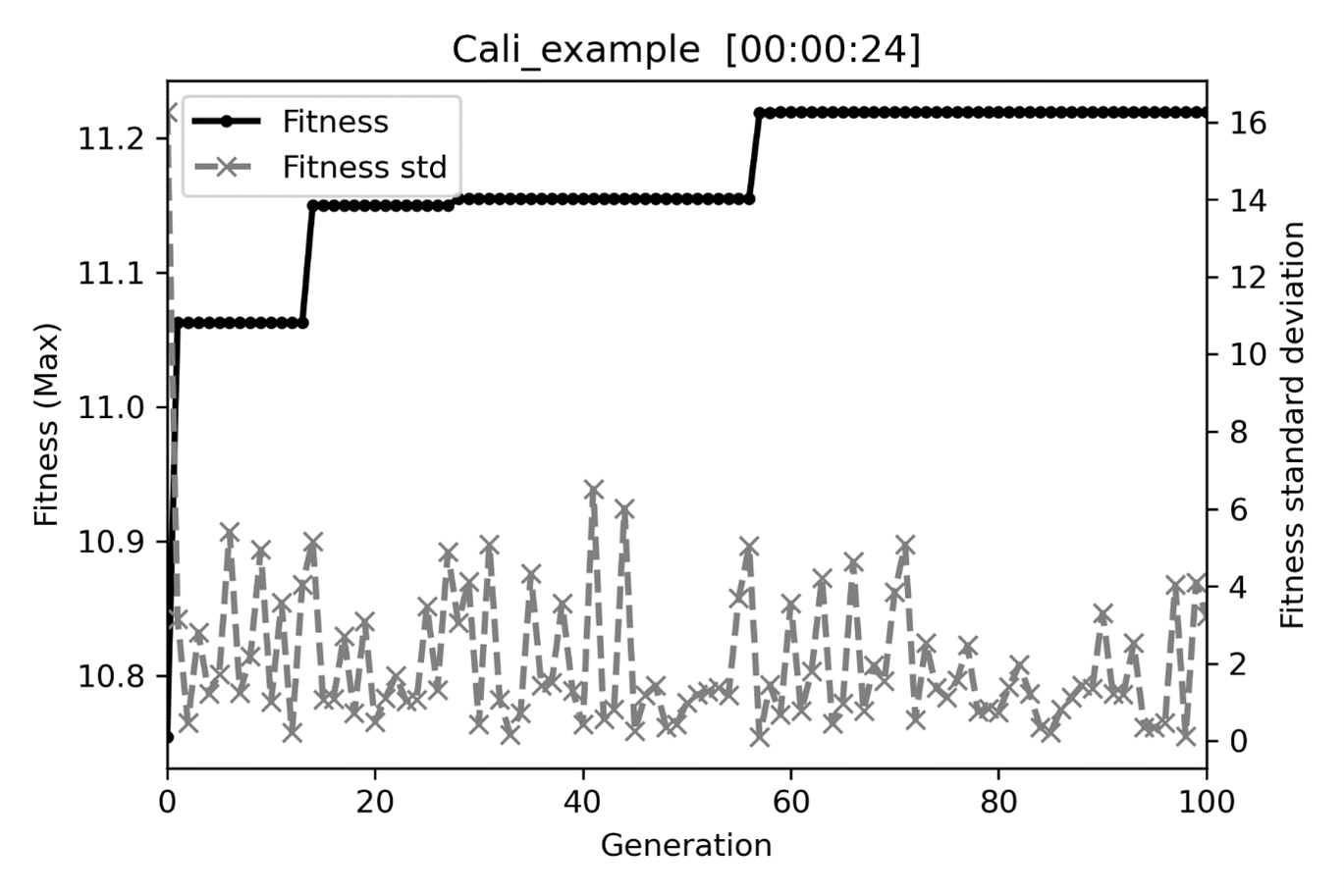
Fig. 7 The fitness and within-population standard deviation plot.
Converter
The Converter is designed to convert a list of parameter DataFrames into a 1-D array for GA calibration and return a 1-D array to the original list of DataFrames. It can automatically exclude NaN values in DataFrames and fix parameters not intended to be calibrated. Also, a converter can generate the calibration input dictionary.
We use the following synthetic list of DataFrames as an example.
import numpy as np
import pandas as pd
import HydroCNHS
import HydroCNHS.calibration as cali
### Prepare testing data.
par_df1 = pd.DataFrame({"Subbasin1": [1000,1000,3], "Subbasin2": [4,5,6]},
index=["a", "b", "c"])
par_df2 = pd.DataFrame({"Agent1": [9,8,7], "Agent2": [6,5,None]},
index=["Par1", "Par2", "Par3"])
bound_df1 = pd.DataFrame({"Subbasin1": [[0,1000],[0,1000],[0,10]], "Subbasin2": [[0,10],[0,10],[0,10]]},
index=["a", "b", "c"])
bound_df2 = pd.DataFrame({"Agent1": [[0,10],[0,10],[0,10]], "Agent2": [[0,10],[0,10],None]},
index=["Par1", "Par2", "Par3"])
df_list = [par_df1, par_df2]
par_bound_df_list = [bound_df1, bound_df2]
Now, we want to generate the calibration input dictionary with fixed “a” and “b” parameters for “Subbasin1.” We can do the following:
### Create an object called Converter.
converter = cali.Convertor()
### Generate GA inputs with fixed a & b parameters for Subbasin1.
fixed_par_list = [[(["a","b"], ["Subbasin1"])], []]
cali_inputs = converter.gen_cali_inputs(
"working directory", df_list, par_bound_df_list, fixed_par_list)
### Get formatter
formatter = converter.formatter
### Show cali_inputs
print(cali_inputs)
r"""
{'wd': 'working directory',
'par_name': ['a|Subbasin2', 'b|Subbasin2', 'c|Subbasin1', 'c|Subbasin2',
'Par1|Agent1', 'Par1|Agent2', 'Par2|Agent1', 'Par2|Agent2',
'Par3|Agent1'],
'par_bound': [[0, 10], [0, 10], [0, 10], [0, 10], [0, 10], [0, 10], [0, 10],
[0, 10], [0, 10]]}
"""
We can see the ‘par_name’ in cali_inputs does not contain ‘a|Subbasin1’ and ‘b|Subbasin1.’ The outputted a formatter contains the relationship between a 1-D array and a list of DataFrames.
Convert from a list of DataFrames to a 1D array
### to 1D array
converter.to_1D_array(df_list, formatter)
r"""
# Out[31]: array([4., 5., 3., 6., 9., 6., 8., 5., 7.])
# Note the order of the array corresponds to "par_name" in the cali_inputs.
"""
Convert from a 1D array to the original list of DataFrames
### to df_list
var_array = np.array([5]*9)
converter.to_df_list(var_array, formatter)
r"""
Out[46]:
[ Subbasin1 Subbasin2
a 1000.0 5.0
b 1000.0 5.0
c 5.0 5.0,
Agent1 Agent2
Par1 5.0 5.0
Par2 5.0 5.0
Par3 5.0 NaN]
"""